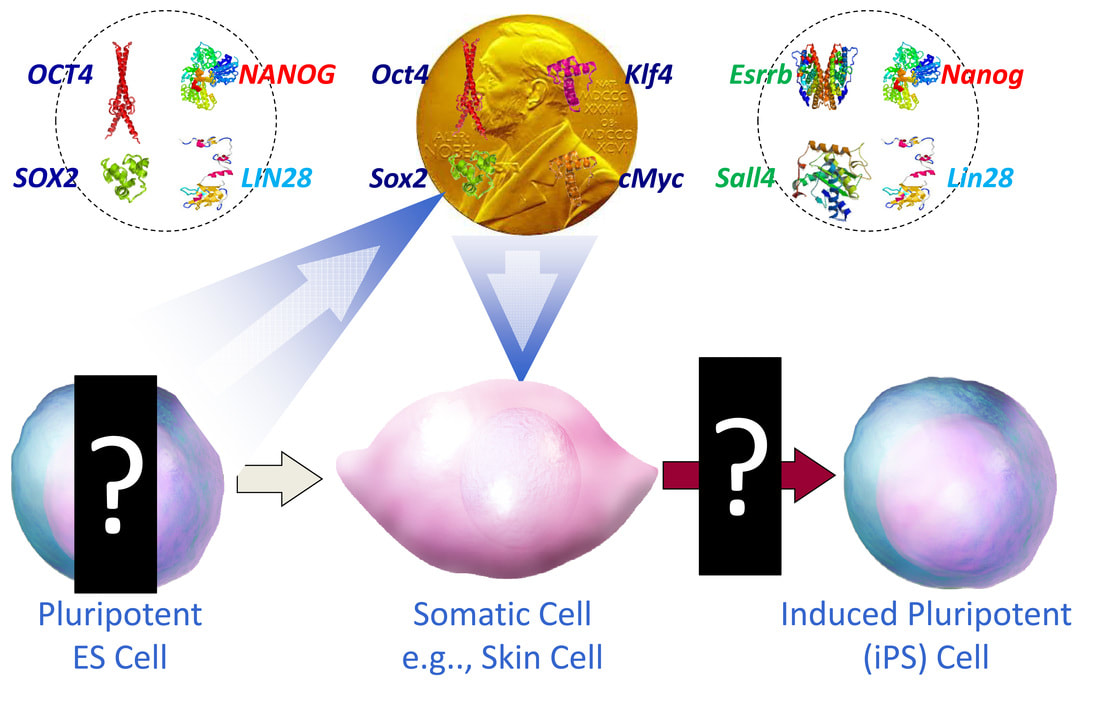
Following fertilization, the totipotent zygote created by fusion of terminally differentiated sperm and oocyte undergoes several cell divisions to give rise to the blastocyst, which is composed of the inner cell mass (ICM) from which pluripotent embryonic stem cells (ESCs) are derived and the trophectoderm that mainly gives rise to the placental tissue. The in vitro capture of pluripotent ESCs under well-defined culture condition for stable maintenance and propagation as well as their potential for regenerative medicine has attracted many including us to study the pluripotency biology. These concerted efforts led to the landmark (Nobel Prize winning) discovery of the generation of induced pluripotent stem cells (iPSCs) in 2006 by Dr. Shinya Yamanaka, who developed an "experimental reprogramming" approach to create iPSCs directly from somatic cells with four transcription factors (Oct4/Pou5f1, Sox2, Klf4 and c-Myc) previously identified as regulators of pluripotency (Cover Figure above).
Dissecting the molecular mechanisms controlling the critical stages of reprogramming (indicated by "?" marks in the Cover Figure above) will lead to discovery of novel reprogramming factors and potential barriers to ground state pluripotency, which will make it possible to more efficiently generate safer iPSCs for therapeutic application. Two major class I histone deacetylase (HDAC)-containing co-repressor complexes, the NuRD and Sin3a complexes, are Nanog partners and both are required at the peri-implantation stage of mouse development, demonstrating the importance of histone deacetylation in cell fate decisions. The NuRD and Sin3a complexes, along with their histone deacetylase activity, are abundant in mammalian cells for transcriptional repression by a number of different sequence-specific transcription factors. Previous investigations by us on the Nanog/Zfp281 partnership led us to the identification of NuRD as a barrier in somatic cell reprogramming (Fidalgo et al., 2011, 2012). Our recent work regarding the Nanog/Tet1 partnership identified Tet1 as an epigenetic regulator that is critical for reprogramming, through its Nanog-dependent transcriptional priming of core pluripotency genes via hydroxymethylation at these loci (Costa, Ding et al., 2013). These studies demonstrate that epigenetic regulators can either positively or negatively modulate Nanog function during the reprogramming process through forming epigenetic modifying complexes and regulating gene transcription. Defining the targets of NuRD-mediated chromatin modification and protein deacetylation, as well as the proteins that target these actions, will allow us to further elucidate the molecular pathways controlling lineage commitment and cell proliferation during mammalian development, and inevitably in cancer. Our lab has been focusing on the following studies to dissect the molecular mechanisms underlying somatic cell reprogramming:
Dissecting the molecular mechanisms controlling the critical stages of reprogramming (indicated by "?" marks in the Cover Figure above) will lead to discovery of novel reprogramming factors and potential barriers to ground state pluripotency, which will make it possible to more efficiently generate safer iPSCs for therapeutic application. Two major class I histone deacetylase (HDAC)-containing co-repressor complexes, the NuRD and Sin3a complexes, are Nanog partners and both are required at the peri-implantation stage of mouse development, demonstrating the importance of histone deacetylation in cell fate decisions. The NuRD and Sin3a complexes, along with their histone deacetylase activity, are abundant in mammalian cells for transcriptional repression by a number of different sequence-specific transcription factors. Previous investigations by us on the Nanog/Zfp281 partnership led us to the identification of NuRD as a barrier in somatic cell reprogramming (Fidalgo et al., 2011, 2012). Our recent work regarding the Nanog/Tet1 partnership identified Tet1 as an epigenetic regulator that is critical for reprogramming, through its Nanog-dependent transcriptional priming of core pluripotency genes via hydroxymethylation at these loci (Costa, Ding et al., 2013). These studies demonstrate that epigenetic regulators can either positively or negatively modulate Nanog function during the reprogramming process through forming epigenetic modifying complexes and regulating gene transcription. Defining the targets of NuRD-mediated chromatin modification and protein deacetylation, as well as the proteins that target these actions, will allow us to further elucidate the molecular pathways controlling lineage commitment and cell proliferation during mammalian development, and inevitably in cancer. Our lab has been focusing on the following studies to dissect the molecular mechanisms underlying somatic cell reprogramming:
1. Zfp281 Function in Somatic Cell Reprogramming
Zfp281 recruits NuRD repressor complex to mediate Nanog autorepression and inhibits somatic cell reprogramming (Fidalgo et al., PNAS 2012). We recently discovered that Zfp281 coordinates opposing functions Tet1 and Tet2 in transcriptional and post-transcriptional control of alternative and/or distinct pluripotency states (Fidalgo et al., Cell Stem Cell 2016). Our ongoing research will further dissect Zfp281 functions in cell lineage specification during early embryo development in both mouse and human.
Zfp281 recruits NuRD repressor complex to mediate Nanog autorepression and inhibits somatic cell reprogramming (Fidalgo et al., PNAS 2012). We recently discovered that Zfp281 coordinates opposing functions Tet1 and Tet2 in transcriptional and post-transcriptional control of alternative and/or distinct pluripotency states (Fidalgo et al., Cell Stem Cell 2016). Our ongoing research will further dissect Zfp281 functions in cell lineage specification during early embryo development in both mouse and human.
Cover Design: DNA methylation and demethylation play a critical role in regulating/reprogramming pluripotent stem cell fates. Fidalgo et al. discovered that the transcription factor Zfp281 partners with Tet methylcytosine dioxygenase 1 (Tet1) in establishing and maintaining the primed pluripotency by antagonizing Tet2 functions that are associated with naive pluripotency. Image by Jill K Gregory, printed with permission from ©Mount Sinai Health System, New York, USA.
2. Tet1 and Tet2 Functions in Somatic Cell Reprogramming
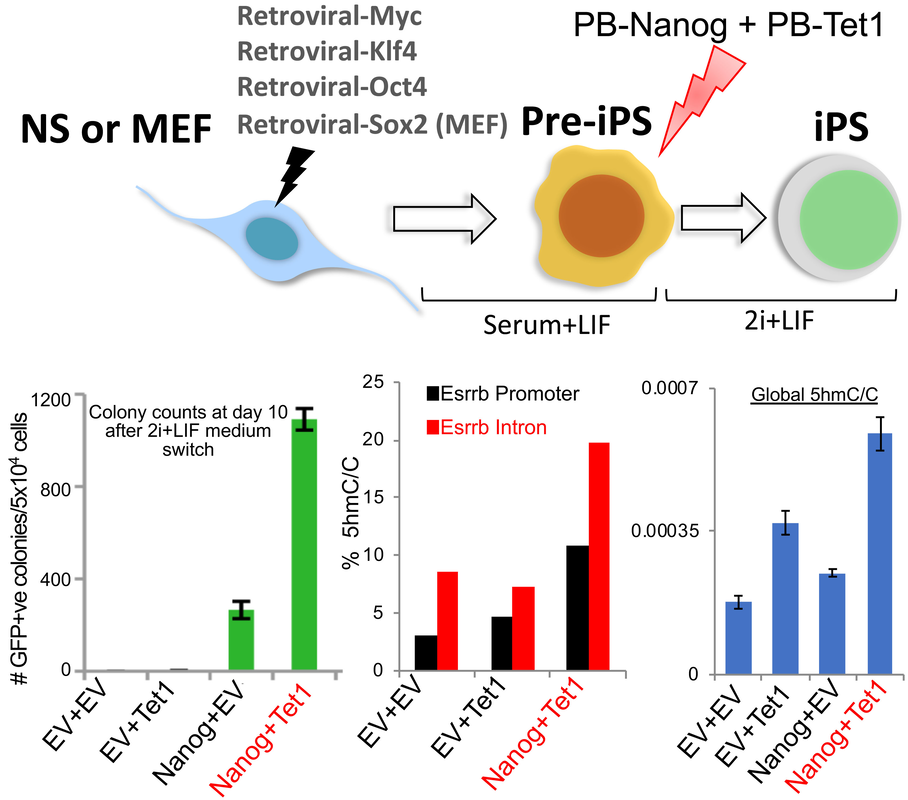
During our studies of an extended Nanog interactome, we have identified many novel protein partners of Nanog, among which are DNA dioxygenases Tet1 and Tet2. We found that, when acting together with Nanog, Tet1 and Tet2 synergistically enhance somatic cell reprogramming in a catalytic-activity dependent manner (Costa, Ding et al., Nature 2013; see illustration below). Our ongoing research focuses on discovering novel Tet1/2 partners and understanding their functions in controlling stem cell pluripotency and somatic cell reprogramming.
During our studies of an extended Nanog interactome, we have identified many novel protein partners of Nanog, among which are DNA dioxygenases Tet1 and Tet2. We found that, when acting together with Nanog, Tet1 and Tet2 synergistically enhance somatic cell reprogramming in a catalytic-activity dependent manner (Costa, Ding et al., Nature 2013; see illustration below). Our ongoing research focuses on discovering novel Tet1/2 partners and understanding their functions in controlling stem cell pluripotency and somatic cell reprogramming.
3. Other Nanog/Oct4 Partners in Pluripotency and Reprogramming Control (ongoing projects)
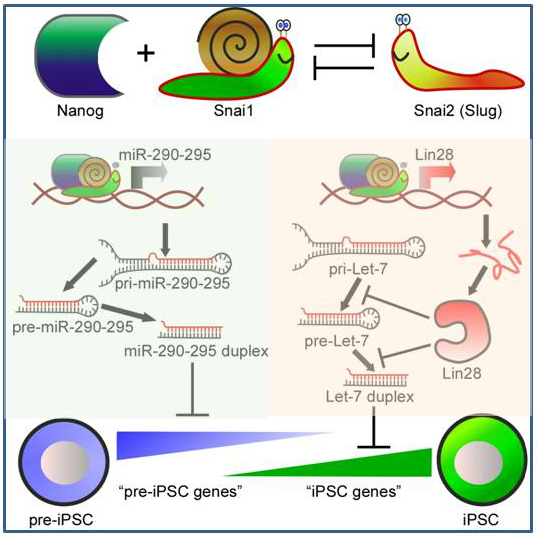
We are in the process of dissecting other novel pluripotency-associated transcription factors and epigenetic regulators from the Nanog/Oct4 interactome for their roles in modulating Nanog/Oct4 function in somatic cell reprogramming. For example, we found that mesenchymal transcription factor Snai1 (aka Snail) can partner with Nanog and facilitate somatic cell reprogramming, and that another mesenchymal transcription factor Snai2 (aka Slug) antagonizes Snail's function by direct transcriptional repression. Such a unique partnership between Nanog and Snail, but not Slug, leads to transcriptional regulation of downstream target genes including miRNAs mediating the final reprogramming outcome (Gingold, Fidalgo et al., Molecular Cell 2014).
We are in the process of dissecting other novel pluripotency-associated transcription factors and epigenetic regulators from the Nanog/Oct4 interactome for their roles in modulating Nanog/Oct4 function in somatic cell reprogramming. For example, we found that mesenchymal transcription factor Snai1 (aka Snail) can partner with Nanog and facilitate somatic cell reprogramming, and that another mesenchymal transcription factor Snai2 (aka Slug) antagonizes Snail's function by direct transcriptional repression. Such a unique partnership between Nanog and Snail, but not Slug, leads to transcriptional regulation of downstream target genes including miRNAs mediating the final reprogramming outcome (Gingold, Fidalgo et al., Molecular Cell 2014).
4. Sox2 and its Partners in Pluripotency and Reprogramming Control.
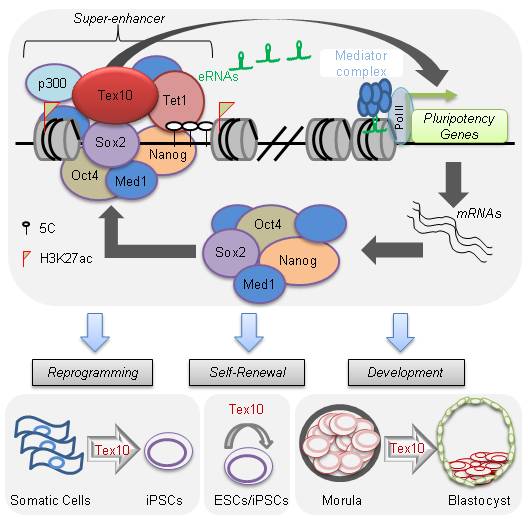
We are interested in understanding how Sox2 guides the enhanceosome assembly and controls the hierarchical transcriptional activation of key pluripotency genes during the last stage of reprogramming. We have recently identified a novel pluripotency factor, namely Tex10, as a close partner of Sox2 that coordinates epigenetic control of super-enhancer activity in pluripotency and reprogramming (Ding et al., Cell Stem Cell 2015).
We are interested in understanding how Sox2 guides the enhanceosome assembly and controls the hierarchical transcriptional activation of key pluripotency genes during the last stage of reprogramming. We have recently identified a novel pluripotency factor, namely Tex10, as a close partner of Sox2 that coordinates epigenetic control of super-enhancer activity in pluripotency and reprogramming (Ding et al., Cell Stem Cell 2015).
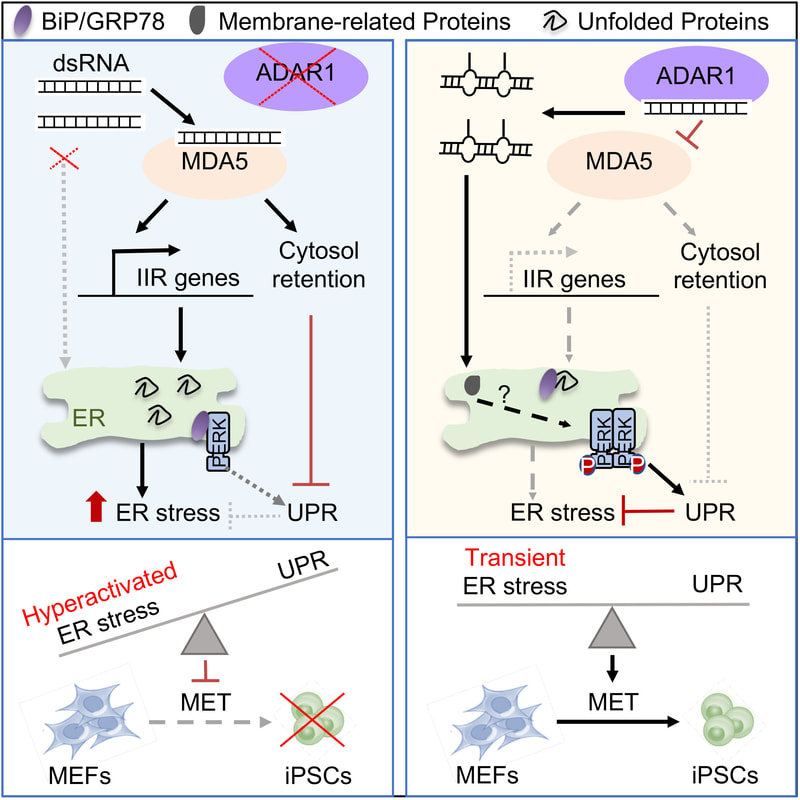
5. ADAR1-dependent RNA editing promotes MET and iPSC reprogramming by alleviating ER stress.
ADAR1-mediated A-to-I deamination of RNA is amongst the most pervasive and prevalent forms of RNA modifications. A-to-I editing can affect not only the coding potential, but also structure and miRNA recognition. Despite its well-recognized important roles in innate immune response and neural development, the role of this epitranscriptomic mark in cellular reprogramming remained unexplored until now. We presented the first comprehensive molecular and functional dissection of A-to-I RNA editing in epithelial cell specification/cellular reprogramming and demonstrated the significance of epitranscriptomic regulation in shaping cellular identity. Specifically, our study establishes a new paradigm in our understanding of cell fate decisions whereby A-to-I RNA editing controls MDA5 sensing of membrane-associated dsRNAs, and by doing so, influences the balance between ER stress/UPR and innate immune response, key cellular processes in development and diseases (Guallar et al., Cell Stem Cell 2020).
ADAR1-mediated A-to-I deamination of RNA is amongst the most pervasive and prevalent forms of RNA modifications. A-to-I editing can affect not only the coding potential, but also structure and miRNA recognition. Despite its well-recognized important roles in innate immune response and neural development, the role of this epitranscriptomic mark in cellular reprogramming remained unexplored until now. We presented the first comprehensive molecular and functional dissection of A-to-I RNA editing in epithelial cell specification/cellular reprogramming and demonstrated the significance of epitranscriptomic regulation in shaping cellular identity. Specifically, our study establishes a new paradigm in our understanding of cell fate decisions whereby A-to-I RNA editing controls MDA5 sensing of membrane-associated dsRNAs, and by doing so, influences the balance between ER stress/UPR and innate immune response, key cellular processes in development and diseases (Guallar et al., Cell Stem Cell 2020).